Yamuna Krishnan is a Professor in the Department of Chemistry at the University of Chicago. In 2002, she completed her PhD in organic chemistry from IISc. Her passion for the exploration of the chemistry of biological systems evolved at the National Centre for Biological Sciences, Bangalore, where she spent eight years running her own lab before moving to Chicago in 2014.
A winner of several accolades, including the Shanti Swarup Bhatnagar Prize and the Infosys Prize, Yamuna’s work is revolutionising our understanding of the environment inside cells. Today, her lab develops molecular tools to study organelles, the membrane-bound compartments found within the cells of eukaryotes (which include fungi, plants, and animals, but not bacteria, which are prokaryotes). Organelles perform specific functions for a cell, much like our organs do for our body. Yamuna has also established two startups that are translating research from her lab to create diagnostic and therapeutic tools.
Yamuna was in IISc on 11 July 2023 for a talk on her work. During her visit, she spoke to CONNECT about her interest in cell biology, research, and her startups.

What led you to choose scientific research as your career path?
When I was a kid, my dad used to always ask me questions about the world around us. If he had a glass with ice cold water, he would ask me why there was frost outside of it. They were very simple things, but they made me learn how to ask questions. He kept that curiosity alive – the education system tends to beat that out of us. My father was an architect, but he encouraged me to think critically. That was probably why I chose a career in science – because I always had this question in my mind: Why are things the way they are?
After studying chemistry, why did you switch to studying cells?
When I was in IISc, I ended up working in Sandhya Visweswariah’s lab [Honorary Professor, Department of Developmental Biology and Genetics] for the very last part of my PhD thesis. Working in her lab was so exciting. She herself was doing experiments with me. The way she ran her lab was very egalitarian, and I liked it so much. That, and seeing how lipids I had made influenced DNA transfection [incorporation of foreign DNA into a genome] – the question I was working on at the time – convinced me that I wanted to do something related to biology. Sandhya was not my PhD advisor, but she has had a great influence on my thinking and my value system. I worked in her lab for a very short time, just six months, but it has had a lifelong effect.
Biology is chemistry that is very complex. When considering molecules and their chemistry, you go from simple systems to complex systems. Either you can go into the complexity of materials that are a mixture of many kinds of chemicals, or you can look at functional complexity inside a complex system, like a biological system. So, for those chemists who don’t want to do “variations on a theme,” it is a natural trajectory to go into either materials or biology.
Biology is chemistry that is very complex
Your lab is known for creating tools to explore the interiors of live cells for their chemical landscapes, and how they change during disease. Why is such an exploration necessary?
We started out by trying to understand how organelle function impacts cell function by mapping the chemical milieu within organelle lumens or cavities. In many diseases, when an organ malfunctions, this malfunction can be traced right down to dysfunctional organelles in the cells comprising that tissue. There is always feedback between organelles and cells. The organelle state and cell state are tightly coupled. If you want to understand how disease affects cells, such new avenues are ripe for exploration.
For example, in heart disease, we know that the Golgi complex is highly fragmented. Biologists and clinicians have known for decades about organelle dysfunction in disease. The reason, I feel, why they have not yet been able to leverage this knowledge, is because our description of an organelle is largely physical and morphological. We say: “It became bigger, or tubular, or crescent-shaped.” But you see, there are molecules that are making this happen. And I believe, as a chemist, that until you get chemical measures and molecular measures of normalcy and abnormality in organelles, we will never be able to understand how organelle dysfunction has affected pathophysiology, and how organelles function in normal physiology.
If we can address such questions, then that might be a way by which we can diagnose, or possibly treat diseases. That’s why my lab developed a platform to quantitatively image chemicals in organelle lumens.
Biologists and clinicians have known for decades about organelle dysfunction in disease
You use nanodevices to study the chemistry of organelles. How do these devices work?
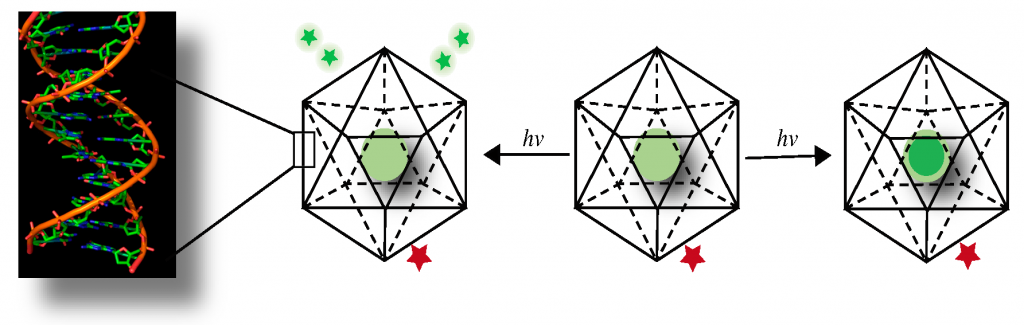
These are DNA-based reporters or probes. If you want to measure an ion inside an organelle, you need a probe that can perform two functions. First, it needs to have a measuring module like an ion-sensitive dye. And second, you need to be able to transport this device inside the target organelle.
Here, we use knowledge from biology. Say you want to target a certain organelle. You identify a shuttle protein that moves between the plasma membrane of the cell and that organelle. If you identify a molecular motif that binds to this shuttle protein and can glue it on to your device, then every single shuttle protein that comes out onto the plasma membrane will grab a DNA device and take it inside. After some time, your target organelle glows brightly because it has become a fancy cuvette containing your DNA sensor from which you can read out the concentration of your ion. With this technology, we can get quantitative chemical information. Here, the DNA probe is just a simple scaffold; it’s like a coat hanger on which you can hang a green dye and a red dye and get a functional readout.
Here is an example. Take a chloride sensitive dye, attach it to one end of your DNA device, and a chloride insensitive dye, and hang it on the other end. Your device will also have a sequence that will help attach it to a shuttle receptor. Simply add it onto the cells, wait for a little time for the cells to internalise it. Wash it off from the surface. Over the next few minutes, the device will move from the surface to the target organelle. By simply imaging the cell using different wavelengths of light, you get a quantity, a number, for the fluorescence of each dye. We can use this to measure the amount of chloride inside that organelle.
A DNA probe is like a coat hanger on which you can hang a green dye and a red dye and get a functional readout
What is the use of the two dyes? How do they help in sensing the changing chemical conditions?
Imagine that I have a constant red light. And then I have a green torch that can sense a certain ion. It is off to start with. As the levels of an ion increase, the intensity of the green light increases. For more amount of the ion, I have more intensity of green for the same amount of red. So, the readout will go from just red, to deep orange, to light orange, to yellow and so on, to lime green and green. That gives me an optical gradient that tells me how much of a certain ion is there. By just looking at green fluorescence becoming brighter or dimmer, my eye cannot tell me the exact concentration. If I have something constant that I can compare it to, then our eyes can gauge the difference. It is the composite that gives you a readout, that gives you a number.
Where have such explorations led?
The organelle state reflects the cell state, and organelle composition defines organelle state. Just like your body temperature, blood pressure, heart rate, and so on are numbers that describe the state of your body, the levels of different ions inside an organelle can describe the organelle state. These numbers will vary when a cell is diseased. By looking at them, one can tell if the cell is healthy or diseased.
One of my companies, Esya Labs, is profiling lysosomes inside cells that are derived from skin biopsies of human patients, and we can diagnose Alzheimer’s disease and many other neurodegenerative diseases way earlier, possibly before patients start manifesting the symptoms of these diseases.

Most organelles are enclosed within a complex network of membranes, and many of them dynamically connect with and separate from each other. Are lysosomes also part of this intracellular membrane network?
Each organelle is a reaction vessel carrying out a different kind of function. The chemical milieu within an organelle has been optimised over evolutionary timescales to promote the specific biochemistry required for its function. For example, the endoplasmic reticulum is creating proteins by joining together amino acids. In the Golgi complex, we are glycosylating – adding sugar chains to – proteins. Lysosomes are the well-known catabolic organelles of the cell. They are like stomachs of the cell, where obsolete or damaged proteins get digested. And yes, lysosomes interact with mitochondria, with endoplasmic reticulum, with plasma membrane – with most organelles.
Lysosome contains calcium inside its lumen. It gives out small spurts of that calcium, which acts like the signal necessary for its fusion with other organelles. This is required when, for example, the plasma membrane breaks, and lysosome goes and fuses at the breakpoint, providing the extra membrane for repair, acting like a band-aid. That’s only one of its functions, there are so many others.
So, DNA nanodevices sense the internal environment of cell organelles. What does this tell us about how cells work?
The devices sense ions. By telling us the levels of ions inside organelles, they tell us about how ion-channels – tunnels through which ions enter and exit – on organelle membranes function, and how this helps the organelle support the cell state. For example, if the cell is in an inflamed state, then the organelles will rise to the occasion – they will remodel, and all of them will function to maintain the cell’s inflamed state.
You have talked about cancerous tumours and how you can make them accessible to the immune system with the help of these tools. Can you tell us a little bit about this line of research?
If you inject our devices into a mouse, they go straight to a certain kind of immune cell called macrophages – white blood cells that eliminate diseased or dying cells, kill pathogens, and stimulate the immune system. Every organ in the body has resident macrophages.
In many types of tumours, nearly 50% of the tumour is made up of macrophages that are called “tumour-associated macrophages.” When you inject the devices into a mouse with such a tumour, they head straight for tumour-associated macrophages. We still don’t understand why they choose these specific macrophages over the ones that are present in any other organs in the body.
You and I are getting cancer every day. Our cells “go bad” all the time. But the immune system identifies these cells and kills them. So, a tumour is basically an indication that the immune system failed to see a cell that “went bad.” Macrophages display fragments of these diseased or “bad cells” on their own surfaces via a process called “antigen presentation.” By doing this, they alert T cells from the immune system to the location of cancer, which then take care of the bad guys.
The tumours that have very bad prognosis have very high numbers of tumour-associated macrophages. These macrophages are very “hungry,” with hyperactive lysosomes. Lysosomes, as you know, degrade substances. So, they gobble up dead or dying “bad cells” within a tumour, and instead of politely snipping them into pieces and presenting them on the surfaces for the T cells to see, they digest the hell out of them all the way down to amino acids, so that they can make new proteins.
If you could somehow send a message to tumour-associated macrophages: “Please show those antigens here,” then the T cells would come in and obliterate the tumour.
So, all we are doing now, is using our DNA nanodevices to send in inhibitors [certain small molecules] of those digestive enzymes that are present inside the lysosomes that chop up tumour cell proteins. These enzymes are called hydrolases. If they are very powerful, poof, everything is cut into amino acids. But if a small molecule binds and gums up these hydrolases, it blunts their activity. So, now the lysosome is not that hungry anymore. It chews up proteins only a little bit and spits out the rest onto the surface, presents the antigens, and now your T cells can come and take care of the tumour.
Thus, using our technology, we slow down the lysosomal action and allow the cancerous cells to become visible to the immune system.
You can send in an inhibitor attached to the DNA device. Is there a limit to the size of the probe that a cell can take in?
Yes, you don’t want to use too much DNA. The more the DNA, the more expensive the therapeutic. You want to have just enough to do the job. That’s why we keep the sizes of these devices to the minimum. We need DNA that is only long enough to keep the two strands together. For example, at room temperature, six base pairs of DNA will melt – that is separate from each other. We just need enough so that at body temperature, the two strands can stay together.
How are these nanodevices protected from degradation by enzymes inside lysosomes?
They are not protected from enzymes inside lysosomes. But remember, every reaction takes time. Every molecule has a half-life. You just work within that half-life. Even the medicines that you take are metabolised inside the body. It’s not like nothing happens to them, right? You eventually excrete them in some other reacted form.
How do you imagine nanotechnology will change our understanding of basic biology and disease treatment in the near future?
This technology has already changed our understanding of basic biology dramatically; so, it is very difficult to predict how it will change. All we can say is that it will change, and change very fast.
In his work Novum Organum, Sir Francis Bacon famously states that “experiments of light,” or discovery, must precede “experiments of fruit,” or those inventions which would follow. As a scientist who has dabbled in both, is there one of these that you find more exhilarating?
“Experiments of light” and “experiments of fruit” are discovery and invention respectively. Two sides of a coin. Biologists are mostly discoverers. They want to ask why things are the way they are. Chemists are basically inventors. We take things of lower value, combine them, and make things of higher value. Both of these are different ways of doing science – discovery and invention. I never partition it and say, “I should do this much of invention and this much of discovery.” It is a creative process, and we must do what needs to be done at the time that it demands to be done.
You mention in several of your interviews and talks from 2012 that you once insisted that you were “not here to do anything useful,” and that you were merely trying to understand how cells work. Today, you are also devising potential diagnostic and treatment methods. What has changed in the last decade that made you revisit your approach?
If you are able to do something – even the most basic research – it may sometimes actually have larger implications. For example, take bacterial immunity. Why would anyone want to know about bacterial immunity, right? But it led to CRISPR-Cas9 [gene editing] technology. Things that seem so basic, that you think they can have no practical application, can sometimes end up with profound applications. So, if you have birthed a particular technology, then it becomes your job to take it to its logical conclusion. You want to have responsible stewardship of anything that you produce. That way you can also actively direct it as a force for good.




